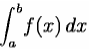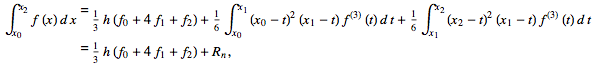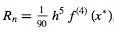Friday, January 13, 2017
Monday, January 25, 2016
Evaluation of Paralympic Basketball Athlete
Protocol of maximum speed in 5 meters starting from the resting state.
Brazilian Paralympic Athlete Paola Klokler (facebook/Paola Klokler 14), Jumper Equipment (facebook/JumperEquipamentos) Athlete.
Wednesday, April 1, 2015
Evaluation in Paralympic Athletics
Angular displacement amplitude of the elbow at the moment of propulsion. Brazilian Paralympic athlete Aline Rocha (facebook/alinerocha), Jumper Equipment (facebook/JumperEquipamentos) athlete.
Thursday, January 1, 2015
Saturday, October 4, 2014
Data Normalization - Body-Size-Independent Measures
Muscle strength is
strongly influenced by body size and correlated with measures such as
Body Mass and Heigth; therefore, utilization of strength
body-size-independent measurements for the aforementioned
applications is important. The importance of this is especially
apparent when comparing persons of different body sizes (ie, athletes
vs nonathletes, men vs women, young vs old), or protocols where Body
Mass could change between data collection periods (eg, long-term
treatment). Normalizing strength measurements to measures of body
size has traditionally been used to remove body-size dependence.
However, there is no consensus on the method by which strength
measurements should be normalized.
While some studies have
not normalized strength measures, others have normalized strength to
Body Mass or a combination of Body Mass and Heigth (ie, Body Mass X
Heigth). Ratio standards (normalizing to Body Mass) assume that
muscle strength is directly proportional to Body Mass. These
assumptions, however, may result in misleading findings. An
alternative to ratio standards is allometric scaling, a normalization
technique based on the theory of “geometric similarity”, or that
the shapes of human bodies are similar, but variable in size.
Allometric scaling is a normalization technique that divides strength
by Body Mass raised to a power that removes body-size effects. The
equation for normalized strength is Sn=S/mb,
where S is the non normalized strength measure, m is Body Mass, b is the allometric b-value
(or scaler), and Sn is the body-size-independent strength
measurement.
In literature exists
generic theoretical reference values for allometric adjustment of
various parameters, but is more appropriate to use the specific
exponent generated for your sample.
The normalization
procedures included plot a linear regression with a log scale of peak
strength parameters of your subjects by your respectives log scale
Body Mass values aand then divide its strength parameters by their
values of body mass raised to the slope found in linear regression.
Here's a link to
download algorithm implemented in Matlab ® to perform the produre to calculate de allometric b-value of your sample. Matlab Code
To use the available
function,first you must create a spreadsheet in excel with the "A"
column containing the body mass in kg of its subjects and the "B"
column containing the respective peaks of the strength parameters of
the subjects. (ie. figure below)
The output of Allometry_gbiomech.m gives you the allometry b-value, the Confidence Interval of the measure and the R Squared value in a dialog box.
References:
- Lleonart, J.; Salat, J.; Torres, G. J. Removing allometric effects of
body size in morphological analysis. Journal of theoretical biology
2000;205:1, p. 85–93.
- Jaric S. Muscle strength testing: use of normalisation for body size. Sports Med 2002;32, p. 615-31.
- Jaric, S.; Mirkov, D.; Markovic, G. Normalizing physical performance tests for body size: a proposal for standardization. Journal of strength and conditioning research / National Strength & Conditioning Association 2005;19:2, p. 467–474.
- Shingleton, A. W. Allometry: The Study of Biological Scaling. Nature Education Knowledge 2010;3:10.
Big Hug!
Big Hug!
Tuesday, August 26, 2014
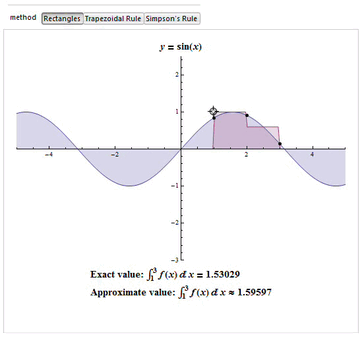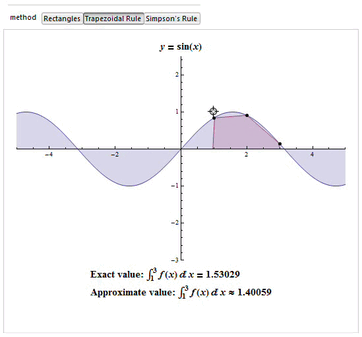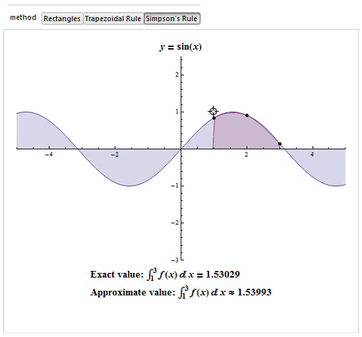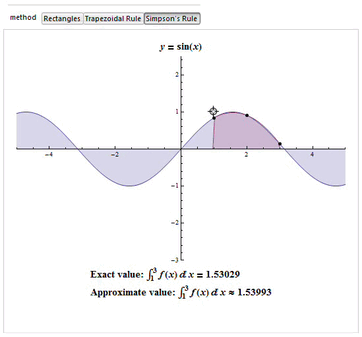
Numerical Integration - Simpson's Rule
 In
numerical analysis, numerical integration constitutes a broad family of
algorithms for calculating the numerical value of a definite integral, and by
extension, the term is also sometimes used to describe the numerical solution
of differential equations. The term numerical quadrature (often abbreviated to
quadrature) is more or less a synonym for numerical integration, especially as
applied to one-dimensional integrals.
In
numerical analysis, numerical integration constitutes a broad family of
algorithms for calculating the numerical value of a definite integral, and by
extension, the term is also sometimes used to describe the numerical solution
of differential equations. The term numerical quadrature (often abbreviated to
quadrature) is more or less a synonym for numerical integration, especially as
applied to one-dimensional integrals.
The
basic problem in numerical integration is to compute an approximate solution to
a definite integral
to a given degree of accuracy. If f(x) is a smooth function integrated over a small number of dimensions, and the domain of integration is bounded, there are many methods for approximating the integral to the desired precision.
An
important part of the analysis of any numerical integration method is to study
the behavior of the approximation error as a function of the number of
integrand evaluations. A method that yields a small error for a small number of
evaluations is usually considered superior. Reducing the number of evaluations
of the integrand reduces the number of arithmetic operations involved, and
therefore reduces the total round-off error. Also, each evaluation takes time,
and the integrand may be arbitrarily complicated.
A
'brute force' kind of numerical integration can be done, if the integrand is
reasonably well-behaved, by evaluating the integrand with very small
increments.
A
large class of quadrature rules can be derived by constructing interpolating
functions that are easy to integrate.
Typically
these interpolating functions are polynomials.
The
simplest method of this type is to let the interpolating function be a constant
function (a polynomial of degree zero) that passes through the point ((a+b)/2,
f((a+b)/2)). This is called the midpoint rule or rectangle rule.
The interpolating function may be an affine function (a polynomial of degree 1) that passes through the points (a, f(a)) and (b, f(b)). This is called the trapezoidal rule.
For either one of these rules, we can make a more accurate approximation by breaking up the interval [a, b] into some number n of subintervals, computing an approximation for each subinterval, then adding up all the results. This is called a composite rule, extended rule, or iterated rule. For example, the composite trapezoidal rule can be stated as
where the subintervals have the form [k h, (k+1) h], with h = (b−a)/n and k = 0, 1, 2, ..., n−1.
Interpolation
with polynomials evaluated at equally spaced points in [a, b] yields the
Newton–Cotes formulas, of which the rectangle rule and the trapezoidal rule are
examples. Simpson's rule is a Newton-Cotes formula for approximating the integral of a function  using quadratic polynomials (i.e., parabolic arcs instead of the straight line segments used in the trapezoidal rule).
using quadratic polynomials (i.e., parabolic arcs instead of the straight line segments used in the trapezoidal rule).
Quadrature
rules with equally spaced points have the very convenient property of
nesting. The corresponding rule with each interval subdivided includes all the
current points, so those integrand values can be re-used.
In sport and exercise biomechanics the data are most commonly available for wich the process of integration is appropiate are data from force platforms and accelerometers. The force data from force plataforms can be used to compute acceleration following Newton's second law (F = m * a). In this example, the process of integration enables the velocity to be obtained, and the process can be repeated in this velocity data in order to obtain displacement. This method can be applied in other type of biomechanics data, for example in EMG, like contractile impulse of activation. Integration of this type of data is best done using numerical integration.
The numerical integration method most commonly found in the literature in the area of biomechanics is the Trapezoidal. However, studies such as Won (2000) shows that we must have concern for the error produced by the choice of the integration method to be used and the method of Simpson is the one with the lowest error.
Simpson's rule can be derived by integrating a third-order Lagrange interpolating polynomial fit to the function at three equally spaced points. In particular, let the function
Since it uses quadratic polynomials to approximate functions, Simpson's rule actually gives exact results when approximating integrals of polynomials up to cubic degree. For example, consider
Then Simpson's rule (which corresponds to the area under the blue curve obtained from the third-order interpolating polynomial) gives
whereas the trapezoidal rule (area under the red curve) gives
with
An extended version of the rule can be written for  tabulated at
tabulated at  ,
,  , ...,
, ...,  as
as
where the remainder term is
for some
Here's a link to download Matlab Code with a sub-program implemented in environment Matlab© to get the integration value of your signal with Simpson's Rule.
To use the function provided just save the file "simpson_gbiomech.m" in the same folder as the data to be analyzed. Then just call in your Matlab routine sub-program as follows in the example below:
Z=simpson_gbiomech(Y); - computes an approximation of the integral of Y via the Simpson's method (with unit spacing). To compute the integral for spacing different from one, multiply Z by the spacing increment.
Z=simpson_gbiomech(X,Y); - computes the integral of Y with respect to X using the Simpson's rule. X and Y must be vectors of the same length, or X must be a column vector and Y an array whose first non-singleton dimension is length(X). SIMPS operates along this dimension.
Note: you can specify the window which will be modified by inserting an index on the input variables.
References:
Big Hug!
To use the function provided just save the file "simpson_gbiomech.m" in the same folder as the data to be analyzed. Then just call in your Matlab routine sub-program as follows in the example below:
Z=simpson_gbiomech(Y); - computes an approximation of the integral of Y via the Simpson's method (with unit spacing). To compute the integral for spacing different from one, multiply Z by the spacing increment.
Z=simpson_gbiomech(X,Y); - computes the integral of Y with respect to X using the Simpson's rule. X and Y must be vectors of the same length, or X must be a column vector and Y an array whose first non-singleton dimension is length(X). SIMPS operates along this dimension.
Note: you can specify the window which will be modified by inserting an index on the input variables.
References:
- Abramowitz, M. and Stegun, I. A. (Eds.). Handbook of Mathematical Functions with Formulas, Graphs, and Mathematical Tables, 9th printing. New York: Dover, p. 886, 1972.
- Davis, P. J.; Rabinowitz, P.. Methods of Numerical Integration.
- Horwitz, A. "A Version of Simpson's Rule for Multiple Integrals." J. Comput. Appl. Math. 134, 1-11, 2001.
- Jeffreys, H. and Jeffreys, B. S. Methods of Mathematical Physics, 3rd ed. Cambridge, England: Cambridge University Press, p. 286, 1988.
- Stoer J.; Bulirsch R.. Introduction to Numerical Analysis. New York: Springer-Verlag, 1980. (See Chapter 3.)
- Trott, M. The Mathematica GuideBook for Programming. New York: Springer-Verlag, p. 105, 2004. http://www.mathematicaguidebooks.org/.
- Whittaker, E. T. and Robinson, G. "The Trapezoidal and Parabolic Rules." The Calculus of Observations: A Treatise on Numerical Mathematics, 4th ed. New York: Dover, pp. 156-158, 1967.
- Won, T. The comparison of among different numerical integration methods on the biomechanics of sport countermovement jump. 18º International Symposium on Biomechanics in Sports. Hong Kong, China: 2000. Available: <https://ojs.ub.uni-konstanz.de/cpa/article/view/2270/2123>
Big Hug!
Subscribe to:
Comments (Atom)